Revolutionizing RNA Research
New Tool Accurately Uncovers the Hidden World of Long-Read Sequencing
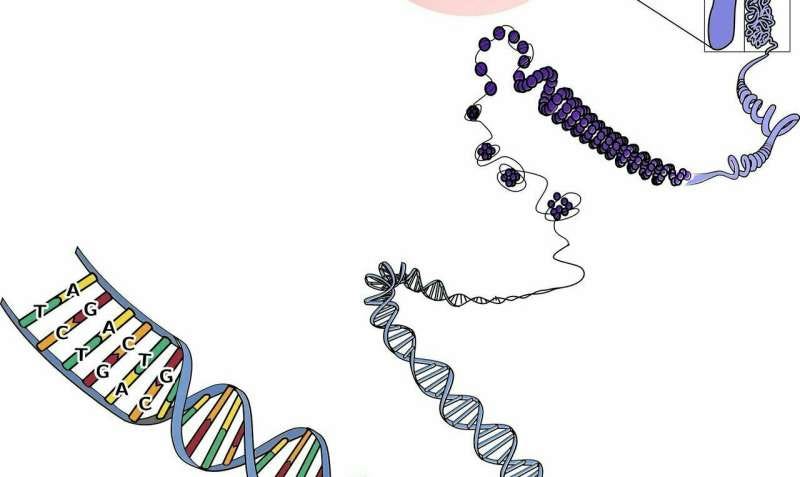
RNA molecules are important for understanding how genes work in the body. They can be cut and joined in different ways, a process called alternative splicing allows a single gene to make several different proteins. This is important in many biological processes like when stem cells turn into specific tissue cells. However, when this process goes wrong, …